MATLAB Deep Learning Toolbox#
Using Ibex GPUs#
Setup#
The intent here is to train on CIFAR10 dataset for 60 Epochs as discussed in here.
The CNN is defined in the M-file and not downloaded from Matlab’s predefined neural network architectures.
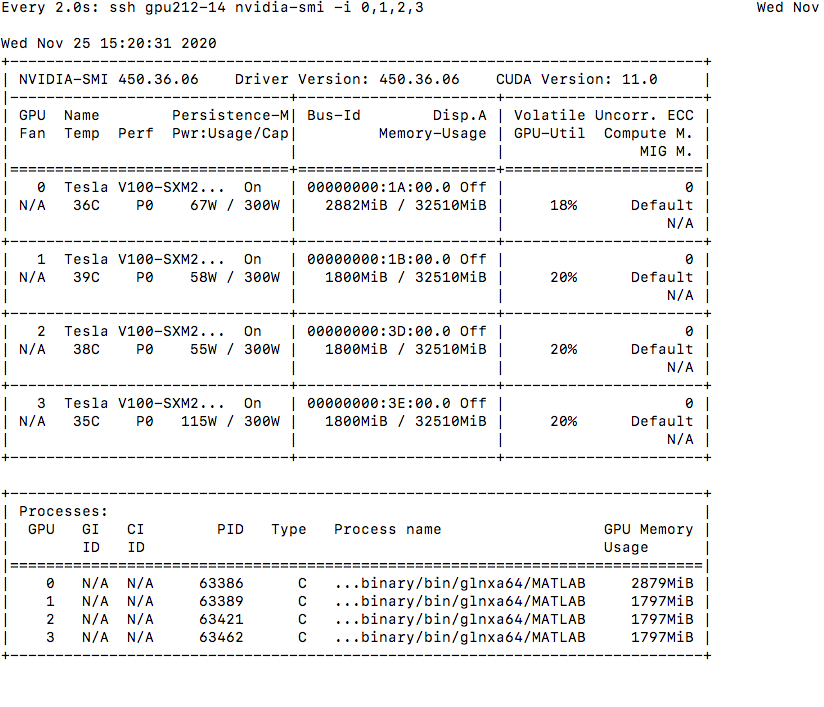
Idea here is to test MultiGPU training. We use 4xV100 nodes with SXM2 (NVLink enabled) on node with SLURM --constraint=gpu_ai added to the jobscript.
The tested version of matlab is R2020a installed on Ibex as a modulefile.
The following files needs to be present in the pwd.
ls *.m
cifar10.m convolutionalBlock.m data_download.m saveCIFAR10AsFolderOfImages.m startup.m
Here is a short explanation of what they contain:
cifar10.m |
This is the main training script. The downloading for data from the web did not work because of the absence of JRE certificate of the website <http://www.cs.toronto.edu/~kriz/cifar.html>_ . Will have an issue with any website without https protocol. I took this part out and have performed this task and creation of dataset folders (Train and Test) separately in |
convolutionalBlock.m |
This contains the function definition |
data_download.m |
This is a two step process. Download tarball manually from here <http://www.cs.toronto.edu/~kriz/cifar-10-matlab.tar.gz>_. Then on run data_download.m on in the current working directory. This should create the required |
saveCIFAR10AsFolderOfImages.m |
This function is referenced in |
startup.m |
This file was required to overwrite the default launching mechanism of mpi processes with system |
startup.m explained#
With the following in cifar10.m, an error is encountered:
numGPUs = 4;
miniBatchSize = 256*numGPUs;
initialLearnRate = 1e-1*miniBatchSize/256;
options = trainingOptions('sgdm', ...
'ExecutionEnvironment','multi-gpu', ... % Turn on automatic multi-gpu support.
'InitialLearnRate',initialLearnRate, ... % Set the initial learning rate.
The error:
>> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> Starting parallel pool (parpool) using the 'local' profile ...
Error using trainNetwork (line 170)
Parallel pool failed to start with the following error. For more detailed
information, validate the profile 'local' in the Cluster Profile Manager.
Caused by:
Error using parallel.Cluster/parpool (line 86)
Parallel pool failed to start with the following error. For more detailed
information, validate the profile 'local' in the Cluster Profile Manager.
Error using parallel.internal.pool.InteractiveClient>iThrowWithCause
(line 670)
Failed to initialize the interactive session.
Error using
parallel.internal.pool.InteractiveClient>iThrowIfBadParallelJobStatus
(line 808)
The interactive communicating job failed with no message.
MATLAB tries to start the processes using its local mpiexec. This is a know issue and the workaround suggested by MATLAB is to create a startup.m file and disable using local mpiexec
Jobscript#
#!/bin/bash
#SBATCH --gres=gpu:4
#SBATCH --constraint=v100,gpu_ai # or any other GPUs, I need 4 because I have num_gpus=4 in cifar10.m
#SBATCH --time=00:30:00
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=4
module load matlab/R2020a
srun -n ${SLURM_NTASKS} -c ${SLURM_CPUS_PER_TASK} matlab -nodisplay -nodesktop -nosplash < cifar10.m
Expected Output#
Loading module for Matlab-R2020a
Matlab-R2020a modules now loaded
< M A T L A B (R) >
Copyright 1984-2020 The MathWorks, Inc.
R2020a Update 3 (9.8.0.1396136) 64-bit (glnxa64)
May 27, 2020
To get started, type doc.
For product information, visit www.mathworks.com.
ans =
logical
1
'downloadCIFARToFolders' is used in the following examples:
Upload Deep Learning Data to the Cloud
Train Network Using Automatic Multi-GPU Support
>> >> >> >>
locationCifar10Train =
'/ibex/scratch/shaima0d/ML_framework_testing/Matlab/testdir/cifar10/cifar10Train'
>>
locationCifar10Test =
'/ibex/scratch/shaima0d/ML_framework_testing/Matlab/testdir/cifar10/cifar10Test'
>> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> >> Starting parallel pool (parpool) using the 'local' profile ...
Connected to the parallel pool (number of workers: 4).
>> >> >> >> >>
accuracy =
0.8918
Expected GPU Utilization#